FDA cleared, software for quantitative analysis of PSMA PET/CT providing standardized, accurate, and rapid total-body quantitative biomarkers for PSMA PET/CT
A deep learning-enabled application
- Automated segmentation and quantification of PSMA uptake in reference organs
- Automated segmentation and localization of PSMA PET lesion candidates
- Automated PSMA quantification at lesion level – tumor (miT), lymph node (miN, miMa), and visceral disease
Why PYLARIFY AI®?
To drive the use of PSMA PET/CT towards better management of prostate cancer patients, quantifying disease burden accurately and reproducibly on a patient level is a requirement. PYLARIFY AI® is the only existing tool to support this in clinical practice.
How does PYLARIFY AI® work?
PYLARIFY AI® automatically analyzes the CT image to segment anatomical regions, including the liver and the thoracic part of the aorta as reference organs. Subsequently, the PET image is analyzed to detect target hotspots, regions of interest (ROIs) having locally elevated PSMA tracer intensities indicative of suspicious tumor tissue and metastasis. ROIs are marked by selecting from predefined hotspots or by manually drawing and are labeled with type and location and quantified in terms of standard uptake values (SUVs) for tracer uptake intensities and their respective volumes. When used by radiology professionals, the standardized PYLARIFY AI report provides consistent and quantitative assessments of PSMA PET/CT.
PYLARIFY AI® Deployment and Integration to fit into the clinical workflow
PYLARIFY AI™ deployment can be facilitated both as a secure web cloud application and as a local server application. The security and patient privacy are paramount to the installation of PYLARIFY AI®.
Once deployed, the adaptive application can be integrated to the institution image acquisition platforms or to the Picture Archiving and Communication Systems (PACS). The report generated from PYLARIFY AI®, in DICOM and PDF format, could be attached in the existing clinical impression for the physician’s review.
Seamless connection with existing clinical workflow (e.g., linking to picture archiving and communication system (PACS) delivers a unique combination of clinical utility in a workflow-friendly package.
PYLARIFY AI is the only FDA-cleared medical device to offer standardized quantitative and accurate reporting of PSMA PET/CT images, including those through the use of PYLARIFY.
Demonstrated value of PYLARIFY AI™

Reproducible and Quantitative disease-burden Indices
- Improved inter-reader reproducibility in staging (κ >0.80) and quantification (ICC: 0.99) of prostate cancer patients2,3
- Quantitative PSMA scan index (PSI) was associated with PSA and Gleason Score2

Accurate Lesion Quantification
- High segmentation and detection accuracy (>90%) for PSMA lesions in regional and distant lymph Nodes.3

High Efficiency
- Significant efficiency in creating quantitative structure reporting. Reader spends an average ~3 min minutes for a comprehensive, quantitative report.2

Clinical Utility
- A quantitative PSMA score/index can help stratify patients for available treatment options2
- A quantitative PSMA score/index can accurately demonstrate disease progression and response2
PYLARIFY AI® is a deep-learning application
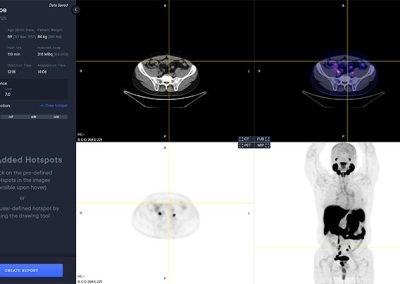
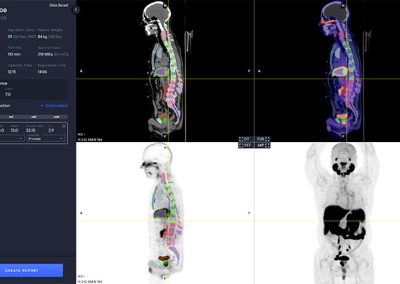
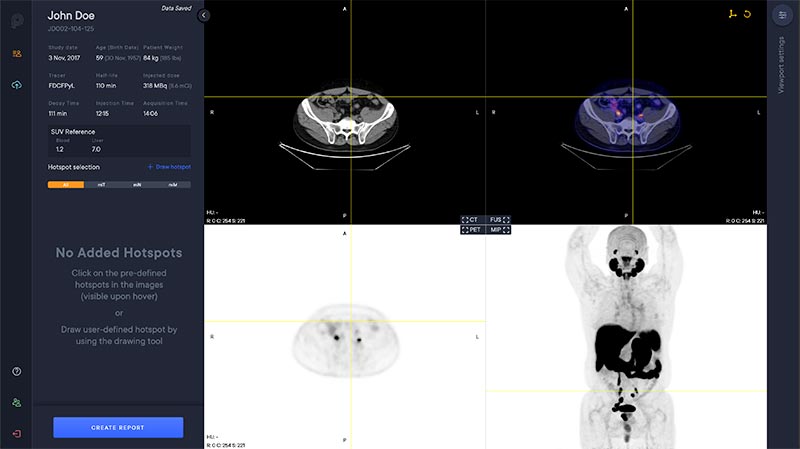
Viewer
The PYLARIFY AI viewer is a multi modal viewer with PET, CT, Fusion and MIP views. The available study information is shown in the control panel to the left.
Segmentation
The software automatically analyzes the CT image to segment anatomical regions, including liver and aorta.
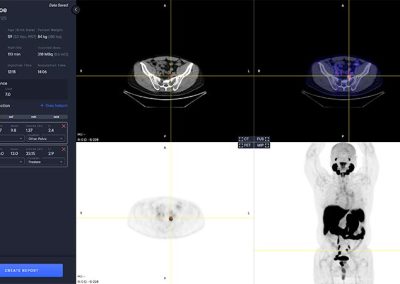
Hotspot Selection and Quantification
Hotspot selection is a tool that enables users to save volumes with high uptake for reporting and quantitative analysis. Each selected hotspot is shown in the left control panel.
References
- Bubendorf, L., et al., Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Human pathology, 2000. 31(5): p. 578-83.
- Pienta KJ, Gorin MA, Rowe SP, Carroll PR, Pouliot F, Probst S, et al. A Phase 2/3 Prospective Multicenter Study of the Diagnostic Accuracy of Prostate Specific Membrane Antigen PET/CT with (18)F-DCFPyL in Prostate Cancer Patients (OSPREY). The Journal of urology. 2021:101097JU0000000000001698.
- Morris MJ, Rowe SP, Gorin MA, Saperstein L, Pouliot F, Josephson D, et al. Diagnostic Performance of (18)F- DCFPyL-PET/CT in Men with Biochemically Recurrent Prostate Cancer: Results from the CONDOR Phase III, Multicenter Study. Clinical cancer research : an official journal of the American Association for Cancer Research. 2021.
- Eiber M, Herrmann K, Calais J, Hadaschik B, Giesel FL, Hartenbach M, et al. Prostate Cancer Molecular Imaging Standardized Evaluation (PROMISE): Proposed miTNM Classification for the Interpretation of PSMA-Ligand PET/CT. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2018;59(3):469-78.
- Fanti S, Minozzi S, Morigi JJ, Giesel F, Ceci F, Uprimny C, et al. Development of standardized image interpretation for 68Ga-PSMA PET/CT to detect prostate cancer recurrent lesions. European journal of nuclear medicine and molecular imaging. 2017;44(10):1622-35.
- Rowe SP, Pienta KJ, Pomper MG, Gorin MA. PSMA-RADS Version 1.0: A Step Towards Standardizing the Interpretation and Reporting of PSMA-targeted PET Imaging Studies. European urology. 2018;73(4):485-7.
- Ceci F, Oprea-Lager DE, Emmett L, Adam JA, Bomanji J, Czernin J, et al. E-PSMA: the EANM standardized reporting guidelines v1.0 for PSMA-PET. European journal of nuclear medicine and molecular imaging. 2021;48(5):1626-38.
- Nickols N, Anand A, Johnsson K, et al. aPROMISE: A Novel Automated-PROMISE platform to Standardize Evaluation of Tumor Burden in (18)F-DCFPyL (PSMA) images of Veterans with Prostate Cancer. J Nucl Med. 2021.
- Johnsson K, Brynolfsson J, Sahlstedt H, et al. Analytical performance of aPROMISE: automated anatomic contextualization, detection, and quantification of [(18)F]DCFPyL (PSMA) imaging for standardized reporting. Eur J Nucl Med Mol Imaging. 2021.