aPROMISE
book a demoIntroducing aPROMISE
aPROMISE is a PACS platform that offers quantitative analysis and standardized reporting of PSMA PET/CT image assessments10. Deep Learning technology on PSMA images to enhance:
- Efficiency: Reduce the laborious task of defining and locating the disease
- Consistency: Enhance the reproducibility and reliability among the readers
- Accuracy: Maintain the high diagnostic accuracy
- Standardization: Enable rapid detection and volumetric quantification of disease burden in PSMA images.
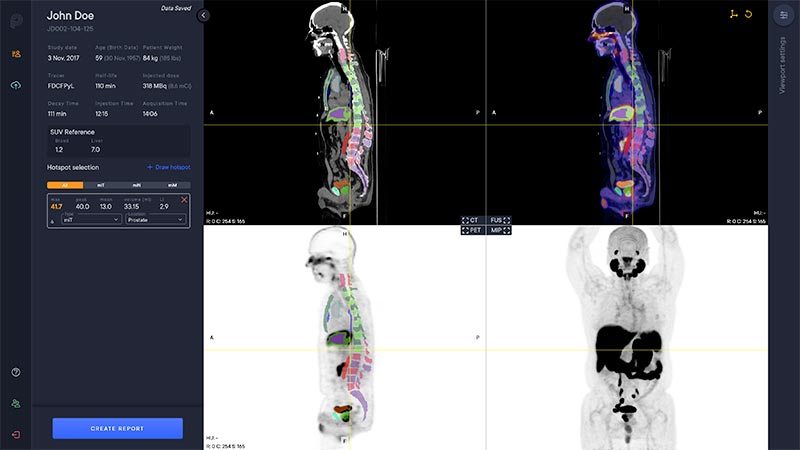
A deep learning-enabled application
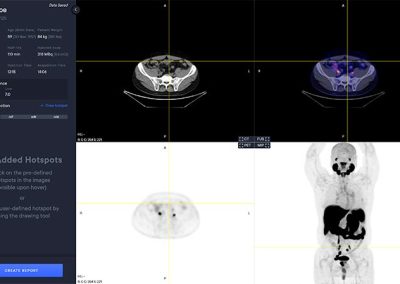
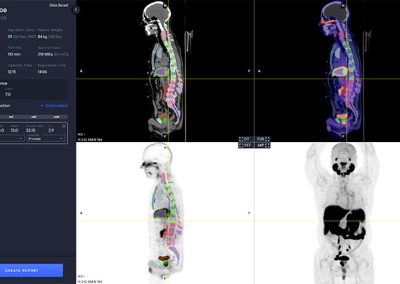
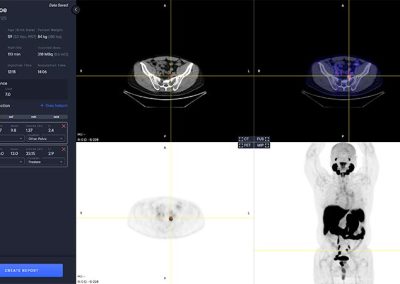
- Automated segmentation and localization of PSMA PET lesion candidates
- Automated segmentation and quantification of PSMA uptake in reference organs
- Automated PSMA quantification at lesion level – tumor (miT), lymph node (miN, miMa), and visceral disease
Interested to learn more? Talk to us!
Through rigorous performance studies, the aPROMISE has demonstrated:

Reproducible and Quantitative disease-burden Indices
- Improved inter-reader reproducibility in staging (κ >0.80) and quantification (ICC: 0.99) of prostate cancer patients2,3
- Quantitative PSMA scan index (PSI) was associated with PSA and Gleason Score2

Accurate Lesion Quantification
- High segmentation and detection accuracy (>90%) for PSMA lesions in regional and distant lymph Nodes.3

High Efficiency
- Significant efficiency in creating quantitative structure reporting. Reader spends an average ~3 min minutes for a comprehensive, quantitative report.2

Clinical Utility
- A quantitative PSMA score/index can help stratify patients for available treatment options2
- A quantitative PSMA score/index can accurately demonstrate disease progression and response2
Legal
Performance and Safety
References
- Bubendorf, L., et al., Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Human pathology, 2000. 31(5): p. 578-83.
- Pienta KJ, Gorin MA, Rowe SP, Carroll PR, Pouliot F, Probst S, et al. A Phase 2/3 Prospective Multicenter Study of the Diagnostic Accuracy of Prostate Specific Membrane Antigen PET/CT with (18)F-DCFPyL in Prostate Cancer Patients (OSPREY). The Journal of urology. 2021:101097JU0000000000001698.
- Morris MJ, Rowe SP, Gorin MA, Saperstein L, Pouliot F, Josephson D, et al. Diagnostic Performance of (18)F- DCFPyL-PET/CT in Men with Biochemically Recurrent Prostate Cancer: Results from the CONDOR Phase III, Multicenter Study. Clinical cancer research : an official journal of the American Association for Cancer Research. 2021.
- Eiber M, Herrmann K, Calais J, Hadaschik B, Giesel FL, Hartenbach M, et al. Prostate Cancer Molecular Imaging Standardized Evaluation (PROMISE): Proposed miTNM Classification for the Interpretation of PSMA-Ligand PET/CT. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2018;59(3):469-78.
- Fanti S, Minozzi S, Morigi JJ, Giesel F, Ceci F, Uprimny C, et al. Development of standardized image interpretation for 68Ga-PSMA PET/CT to detect prostate cancer recurrent lesions. European journal of nuclear medicine and molecular imaging. 2017;44(10):1622-35.
- Rowe SP, Pienta KJ, Pomper MG, Gorin MA. PSMA-RADS Version 1.0: A Step Towards Standardizing the Interpretation and Reporting of PSMA-targeted PET Imaging Studies. European urology. 2018;73(4):485-7.
- Ceci F, Oprea-Lager DE, Emmett L, Adam JA, Bomanji J, Czernin J, et al. E-PSMA: the EANM standardized reporting guidelines v1.0 for PSMA-PET. European journal of nuclear medicine and molecular imaging. 2021;48(5):1626-38.
- Nickols N, Anand A, Johnsson K, et al. aPROMISE: A Novel Automated-PROMISE platform to Standardize Evaluation of Tumor Burden in (18)F-DCFPyL (PSMA) images of Veterans with Prostate Cancer. J Nucl Med. 2021.
- Johnsson K, Brynolfsson J, Sahlstedt H, et al. Analytical performance of aPROMISE: automated anatomic contextualization, detection, and quantification of [(18)F]DCFPyL (PSMA) imaging for standardized reporting. Eur J Nucl Med Mol Imaging. 2021.
- A unique configuration is also available in EU under the name PYLCLARI AI